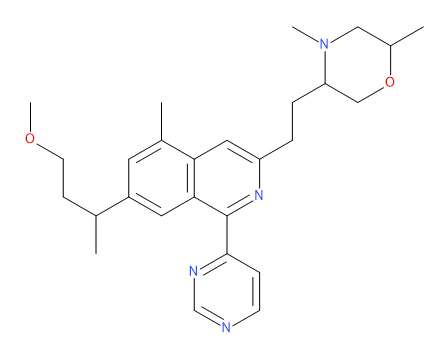
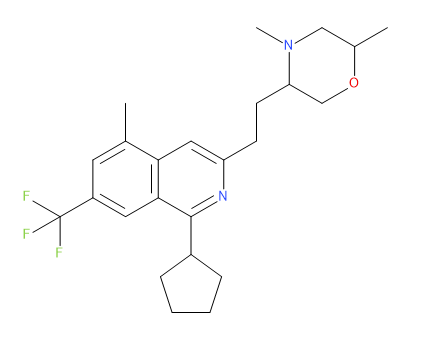
Rgroup Enumeration | |||||
|
| ||||
The Rgroup enumeration generation method requires a core molecule (MOL file) with numbered R atoms specifying the attachment points, and sets of fragments (SD files) to connect to each attachment point. Each fragment must contain one R atom that specifies the position where the fragment is connected to the core molecule.
In the following example, the structure represents a core molecule with two attachment points, R1 and R2:

The set of fragments to be attached at positions R1 and R2 can include any structure that contains a single R atom. For example, for R1 you could specify small chains that include an R atom:



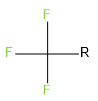
Then, for R2 you could specify single ring systems that include an R atom:
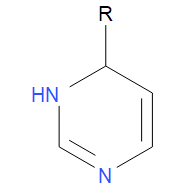


The Rgroup enumeration method then generates molecules by selecting fragments from each dataset stochastically and connecting them to the attachment points. For example: